Marked in green is the current mutation location in the central moving average plot of the normalized frequencies of activating mutations in human kinases using EGFR_HUMAN as the reference sequence.
First, a multiple sequence alignment for the tyrosine kinase domain was obtained from Pfam and additional refinements were done using Muscle. All mutations were then mapped onto the MSA as can be seen here
Second, we computationally calculated a relative frequency for each mutation by dividing the number of samples carrying the mutation by the total number of samples where the corresponding gene has been sequenced (according to COSMIC) and then multiplying the resulting figure by 1,000. The graph below was constructed using the relative frequency of all mutations. For each position of the MSA an accumulated relative frequency was computed by adding the individual relative frequencies of all activating mutations in all human kinases affecting that column of the alignment and a normalized frequency of activating mutations per column was obtained using the formula:
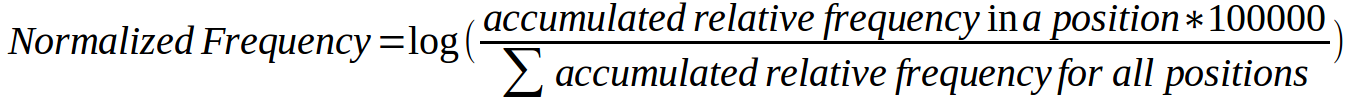
Normalized frequencies were used to calculate central moving averages with a window size of n=13. Values per position of the moving average window are plotted below.
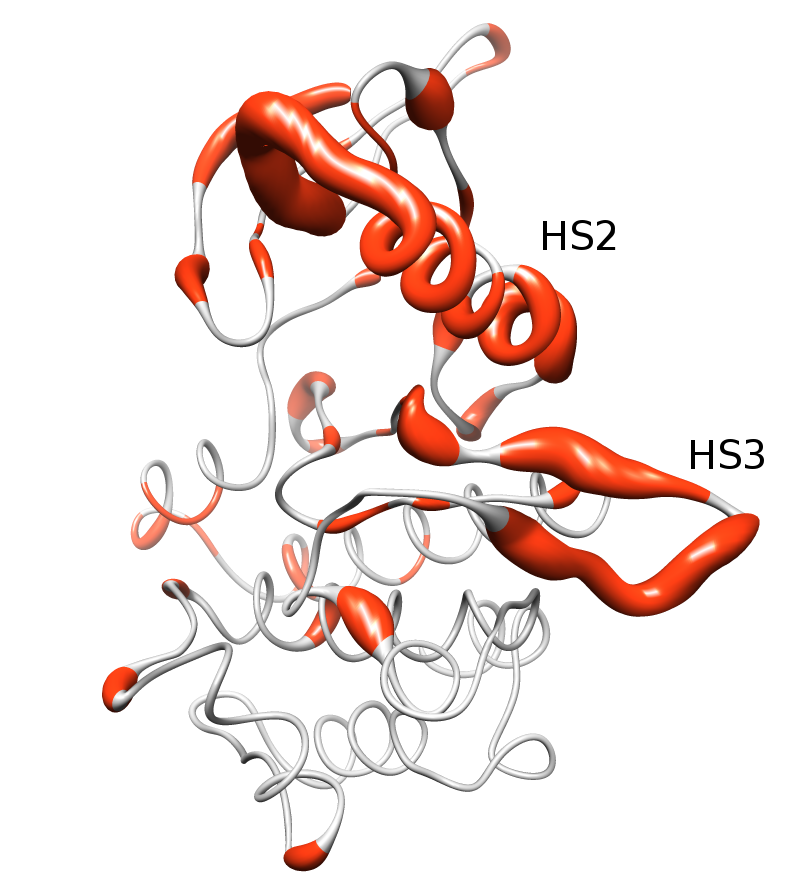 | 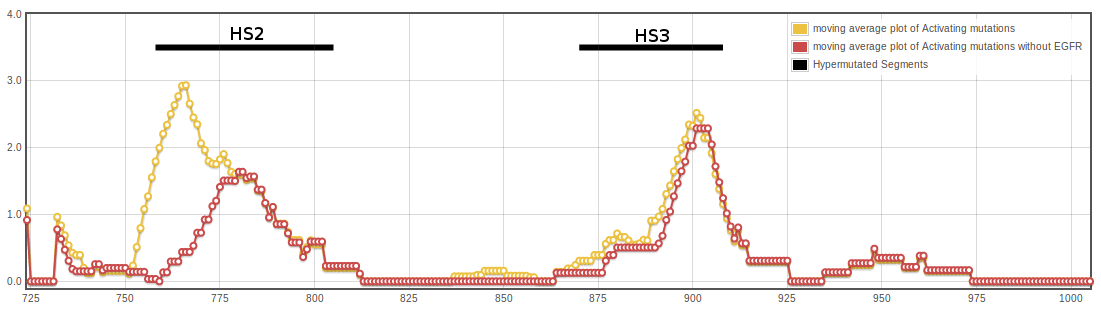 |
Activating mutations cluster in two hypermutated segments of the kinase domain. The image above show the distribution of mutations in the kinase domain of Tyrosin Kinases, depicted in the ribbon representation of the human EGFR structure (pdb code 1M14). Mutated positions are shown in orange, and the diameter of the ribbon is proportional to the normalized frequency of mutations affecting each position.
For more information, please refer to Molina-Vila et al, 2014
| PDB id | Description | Resolution | SPACI Score |
| 3POZ_A | egfr kinase domain complexed with tak-285 | 1.5 | 0.598043 |
| 2RGP_A | structure of egfr in complex with hydrazone, a potent dual inhibitor | 2 | 0.398807 |
| 3VJN_A | crystal structure of the mutated egfr kinase domain (g719s/t complex with amppnp. | 2.34 | 0.357415 |
| 1XKK_A | egfr kinase domain complexed with a quinazoline inhibitor- gw572016 | 2.4 | 0.349657 |
| 3BEL_A | x-ray structure of egfr in complex with oxime inhibitor | 2.3 | 0.334124 |
| 3UG2_A | crystal structure of the mutated egfr kinase domain (g719s/t complex with gefitinib | 2.5 | 0.32858 |
| 2EB2_A | crystal structure of mutated egfr kinase domain (g719s) | 2.5 | 0.326237 |
| 2ITV_A | crystal structure of egfr kinase domain l858r mutation in complex with amp-pnp | 2.47 | 0.319794 |
| 2ITN_A | crystal structure of egfr kinase domain g719s mutation in complex with amp-pnp | 2.47 | 0.319681 |
| 3VJO_A | crystal structure of the wild-type egfr kinase domain in com amppnp. | 2.64 | 0.304627 |
| 2GS6_A | crystal structure of the active egfr kinase domain in complex with an atp analog-peptide conjugate | 2.6 | 0.296067 |
| 4G5J_A | crystal structure of egfr kinase in complex with bibw2992 | 2.8 | 0.292509 |
| 2ITQ_A | crystal structure of egfr kinase domain g719s mutation in complex with afn941 | 2.68 | 0.282167 |
| 2EB3_A | crystal structure of mutated egfr kinase domain (l858r) in c with amppnp | 2.84 | 0.281051 |
| 4HJO_A | crystal structure of the inactive egfr tyrosine kinase domai erlotinib | 2.75 | 0.277641 |
| 2ITZ_A | crystal structure of egfr kinase domain l858r mutation in complex with iressa | 2.72 | 0.273453 |
| 2ITP_A | crystal structure of egfr kinase domain g719s mutation in complex with aee788 | 2.74 | 0.27047 |
| 2GS2_A | crystal structure of the active egfr kinase domain | 2.8 | 0.267665 |
| 3GOP_A | crystal structure of the egf receptor juxtamembrane and kina | 2.8 | 0.267089 |
| 2GS7_A | crystal structure of the inactive egfr kinase domain in complex with amp-pnp | 2.6 | 0.265955 |
| 2GS7_B | crystal structure of the inactive egfr kinase domain in complex with amp-pnp | 2.6 | 0.265955 |
| 2ITW_A | crystal structure of egfr kinase domain in complex with afn941 | 2.88 | 0.264271 |
| 1M14_A | tyrosine kinase domain from epidermal growth factor receptor | 2.6 | 0.262527 |
| 3UG1_A | crystal structure of the mutated egfr kinase domain (g719s/t the apo form | 2.75 | 0.262309 |
| 2ITT_A | crystal structure of egfr kinase domain l858r mutation in complex with aee788 | 2.73 | 0.261026 |
| 2ITU_A | crystal structure of egfr kinase domain l858r mutation in complex with afn941 | 2.8 | 0.258994 |
| 3LZB_B | egfr kinase domain complexed with an imidazo[2,1-b]thiazole | 2.7 | 0.258584 |
| 3LZB_C | egfr kinase domain complexed with an imidazo[2,1-b]thiazole | 2.7 | 0.258584 |
| 3LZB_D | egfr kinase domain complexed with an imidazo[2,1-b]thiazole | 2.7 | 0.258584 |
| 3LZB_A | egfr kinase domain complexed with an imidazo[2,1-b]thiazole | 2.7 | 0.258584 |
| 1M17_A | epidermal growth factor receptor tyrosine kinase domain with 4-anilinoquinazoline inhibitor erlotinib | 2.6 | 0.249629 |
| 2ITX_A | crystal structure of egfr kinase domain in complex with amp- pnp | 2.98 | 0.24909 |
| 2J5F_A | crystal structure of egfr kinase domain in complex with an irreversible inhibitor 34-jab | 3 | 0.242153 |
| 3GT8_A | crystal structure of the inactive egfr kinase domain in complex with amp-pnp | 2.96 | 0.225998 |
| 3GT8_B | crystal structure of the inactive egfr kinase domain in complex with amp-pnp | 2.96 | 0.225998 |
| 3GT8_D | crystal structure of the inactive egfr kinase domain in complex with amp-pnp | 2.96 | 0.225998 |
| 3IKA_A | crystal structure of egfr 696-1022 t790m mutant covalently binding to wz4002 | 2.9 | 0.224373 |
| 2RFE_A | crystal structure of the complex between the egfr kinase domain and a mig6 peptide | 2.9 | 0.22286 |
| 2RFE_D | crystal structure of the complex between the egfr kinase domain and a mig6 peptide | 2.9 | 0.22286 |
| 2JIT_B | crystal structure of egfr kinase domain t790m mutation | 3.1 | 0.216522 |
| 2J5E_A | crystal structure of egfr kinase domain in complex with an irreversible inhibitor 13-jab | 3.1 | 0.21627 |
| 2J6M_A | crystal structure of egfr kinase domain in complex with aee788 | 3.1 | 0.209579 |
| 2JIU_B | crystal structure of egfr kinase domain t790m mutation in complex with aee788 | 3.05 | 0.198567 |
| 2ITO_A | crystal structure of egfr kinase domain g719s mutation in complex with iressa | 3.25 | 0.193627 |
| 4G5P_A | crystal structure of egfr kinase t790m in complex with bibw2 | 3.17 | 0.181471 |
| 4G5P_B | crystal structure of egfr kinase t790m in complex with bibw2 | 3.17 | 0.181471 |
| 2ITY_A | crystal structure of egfr kinase domain in complex with iressa | 3.42 | 0.177366 |
| 2RFD_A | crystal structure of the complex between the egfr kinase domain and a mig6 peptide | 3.6 | 0.133577 |
| 2RF9_A | crystal structure of the complex between the egfr kinase domain and a mig6 peptide | 3.5 | 0.0714469 |
| 2JIV_A | crystal structure of egfr kinase domain t790m mutation in compex with hki-272 | 3.5 | 0.0126281 |
| 2JIV_B | crystal structure of egfr kinase domain t790m mutation in compex with hki-272 | 3.5 | 0.0126281 |
| 4I1Z_A | |||
| 4I20_A | |||
| 4I21_A | |||
| 4I21_B | |||
| 4I22_A | |||
| 4I23_A | |||
| 4I24_A | |||
| 3W2O_A | |||
| 4I24_B | |||
| 3W2P_A | |||
| 3W2Q_A | |||
| 3W2R_A | |||
| 3W2S_A |
|